Week1-16
Cell culture and construction sorafenib-resistant cell line.
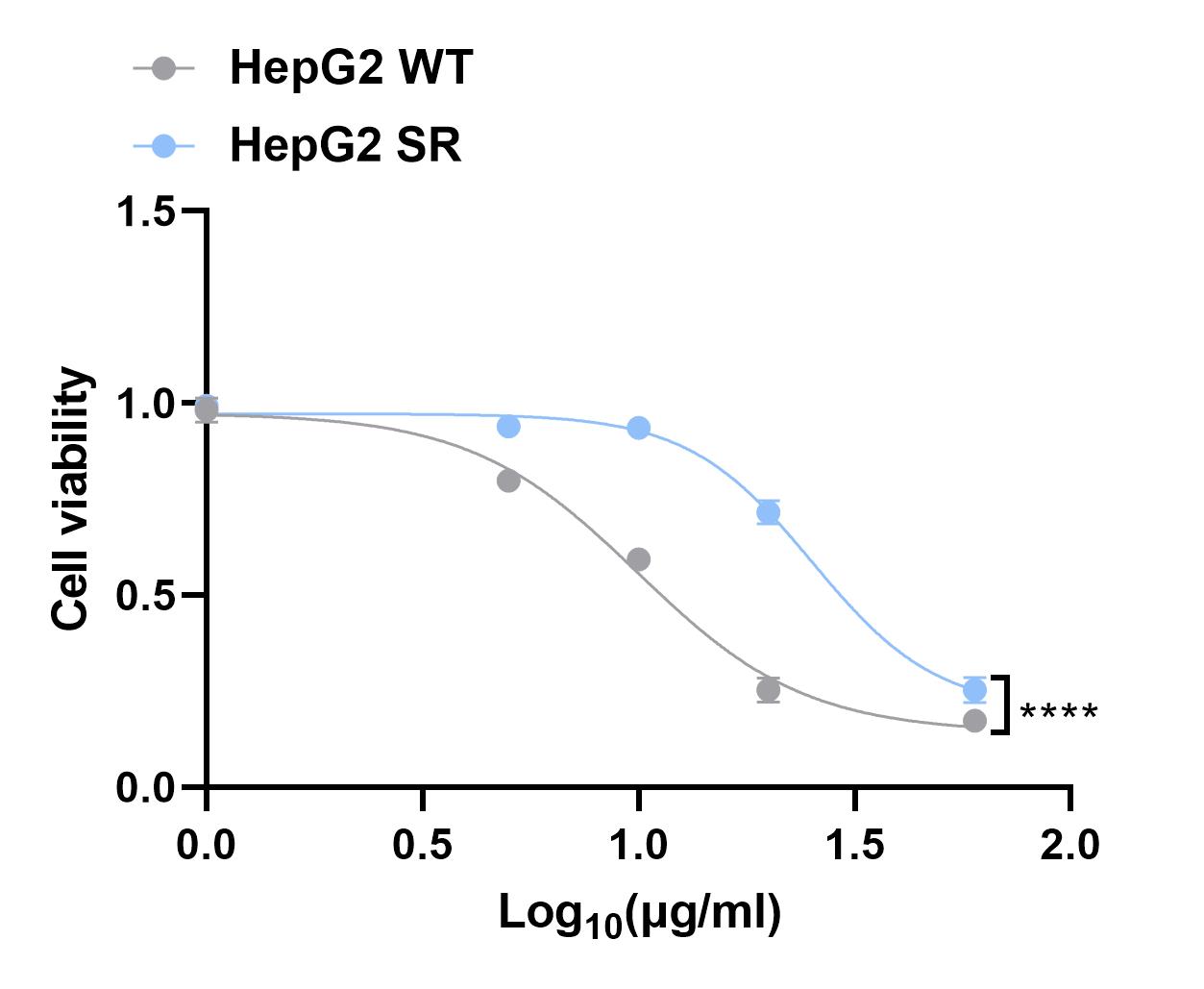
Cells were treated with 2.5 μM sorafenib as the initial concentration, each increase was 0.5μM when the cells tolerated. The final concentration is 7μM and the cells can proliferate normally.Gradually to 0.5 uM drug resistant cell line HepG2 SR can survive in 7uM drug medium for 1 weeks and stable proliferation.
Next, we used CCK8 assay to detect the IC50 of HepG2 SR and HepG2 WT cells.
Week 17-20
RNA sequencing of HepG2 SR and control cells was performed.
Week 21
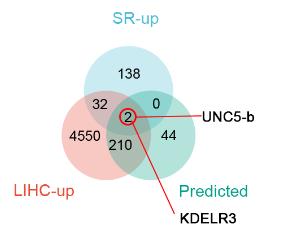
Performing analysis of RNA sequencing results and selecting target molecule.
After obtaining the data, we performed the analysis.Upregulated genes in HepG2 SR, genes upregulated in hepatocellular carcinoma in TCGA database and transcriptomic data based on prediction of liver cancer (TCGA-LIHC) patients in TCGA.
Week22-24
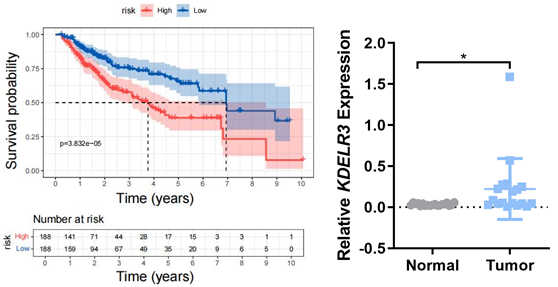
Collecting clinical samples from patients with hepatocellular carcinoma, analyzed the survival period of patients with different KDELR3 contents, and compared the KDELR3 content in tumor cells and normal cells.
Using TCGA to analysis the overall survival for KDELR3 in LIHC and collecting HCC samples to detect the expression of KDELR3.
Week25
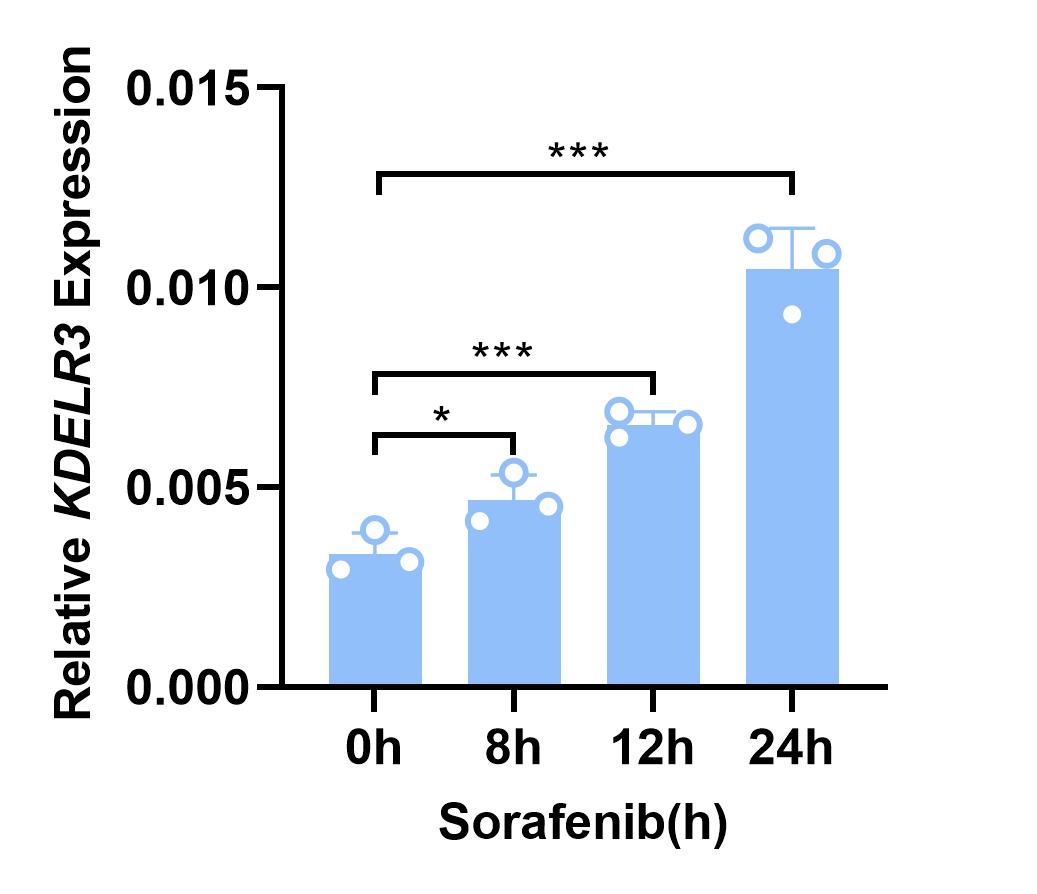
KDELR3 expression was determined in Cells stimulated with sorafenib. The experiment was repeated twice.
Week 26-27
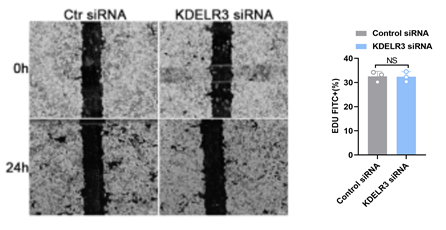
We knocked down KDELR3 expression by siRNA and examined tumor cell proliferation,cell death and migration in HCC cells by using Edu assay, Flow cytometer ,and Scratch assay,. Finally, we analyzed the data and summarized the conclusions.
Week 27-33
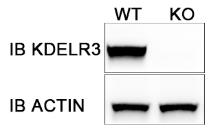
We constructed HepG2 KDELR3 KO cell line by CRISPR/Cas9 technique. During 27-29 week,we constructed KDELR3-Cas9 plasmid. Then we transfected it into HepG2 cell line and performe cell sorting.The monoclones were cultured for two weeks . WB experiment showed that KDELR3 knockout cell line was successfully constructed.
Week 33-41
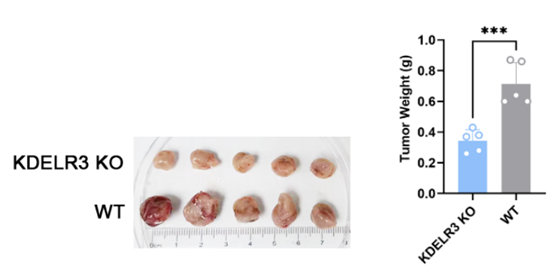
After HepG2 KDELR3 KO cell line were constructed, WT and KO cells were implanted subcutaneously in nude mice. The tumor was monitored and sorafenib was given when the tumor volume reached 150-200mm3(30mg/kg, oral gavage once every two days). Tumor size was monitored by calipers for every 5 days, and the tumor volumes were calculated using the equation (larger diameter) × (smaller diameter)2/2.
Week 33-37
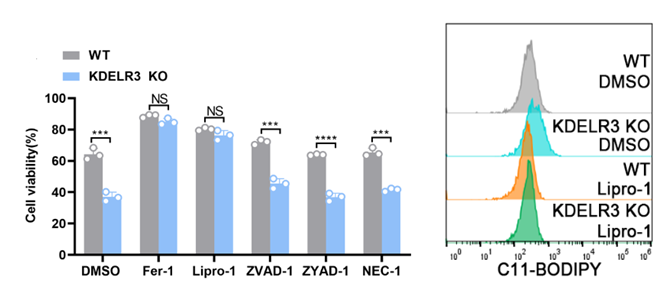
Transcriptome sequencing and KEGG pathway enrichment analysis were performed on WT and KDELR3 KO cells, and ferroptosis pathway was significantly enriched. To verify the sequencing results, we used different programmed death (PCD) inhibitors to pretreat the cells.
Lipid peroxidation is a sign of ferroptosis.The degree of lipid peroxidation of cell membrane was detected by BODIPYTM 581/591 C11 fluorescent dye. We found that the ferroptosis inhibitor can decrease significantly the degree of PCD in KO group .This means that ferroptosis inhibitor had a certain remedial effect and KDELR3 works by preventing sorafenib from inducing ferroptosis .
We found that the degree of lipid peroxidation of cell membrane in KO group was significantly higher than that in control group (DMSO).The addition of ferroptosis inhibitor can effectively reduce this lipid peroxidation.
Week 37-41
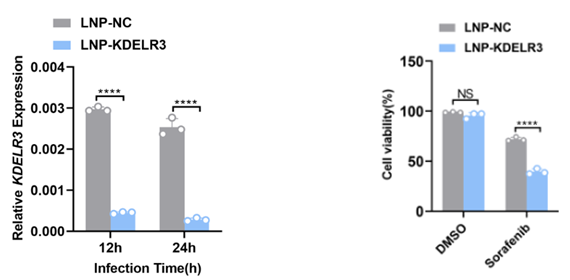
We use LNP to encapsulate the siRNA of KDELR3 and test the encapsulation efficiency and interference efficiency. The results showed that LNP-KDELR3 could decrease KDELR3 expression significantly. Then we pretreated the cells with LNP-KDELR3 and then stimulated them with sorafenib. Compared with the control group, we found that the cell activity of LNP-KDELR3 group decreased significantly after sorafenib stimulation.
 ProjectDesign
ProjectDesign